A web server to approximate the sequence of a generic protein from an in silico library of translatable G-quadruplex (TG4)-mapped peptides
NOTE:
|
Contact information:
Siddhartha Kundu, MD, PhD
Associate Professor,
Department of Biochemistry,
All India Institute of Medical Sciences,
Ansari Nagar, New Delhi - 110029, INDIA
Email:
sidddhartha_kundu@yahoo.co.in siddhartha_kundu@aiims.edu
Siddhartha Kundu, MD, PhD
Associate Professor,
Department of Biochemistry,
All India Institute of Medical Sciences,
Ansari Nagar, New Delhi - 110029, INDIA
Email:
sidddhartha_kundu@yahoo.co.in siddhartha_kundu@aiims.edu
| Schema to delineate peptides that correspond to TG4 | |
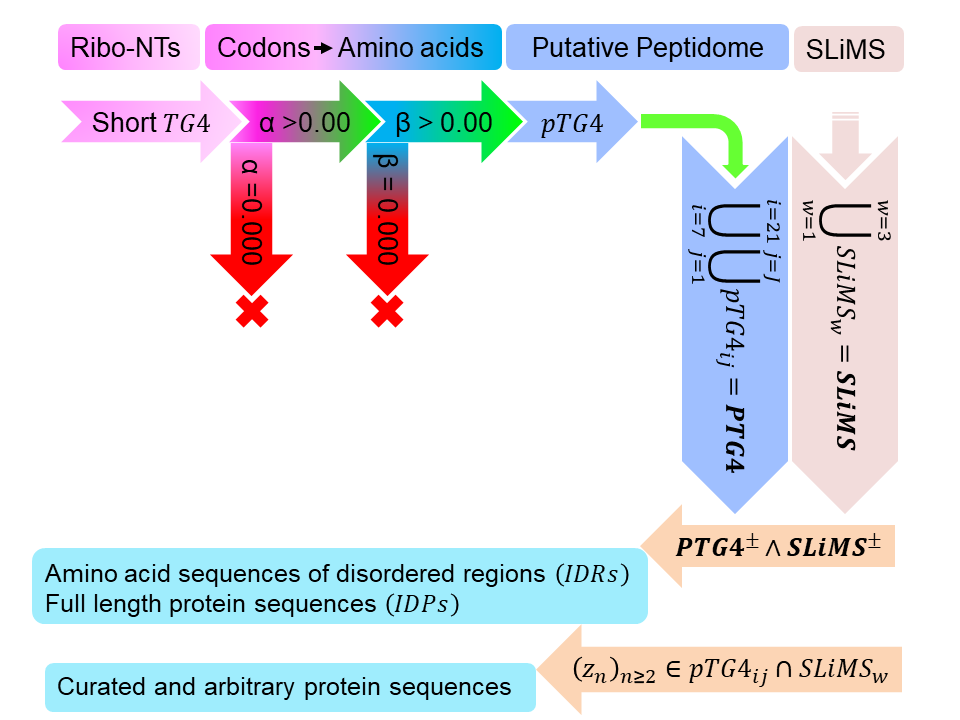 | Algorithm/pipeline of back-end analysis [1] |
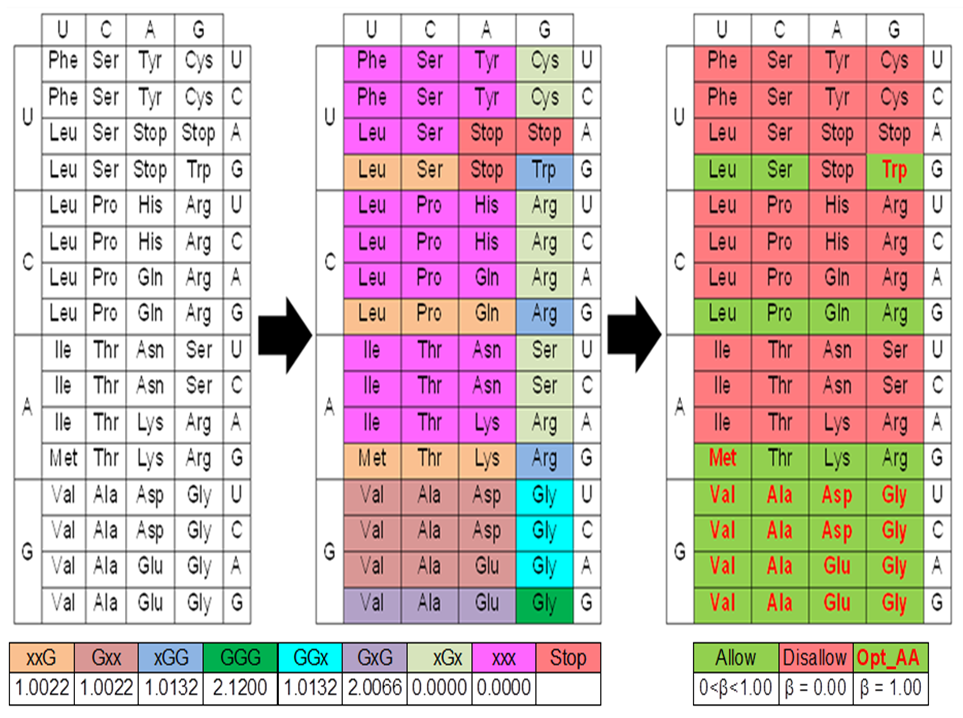 | Codon table to identify suitable amino acids for peptide model of TG4 (~PTG4) [1] |
|
CITATION: [1] Kundu S. Mathematical model of a short translatable G-quadruplex and an assessment of its relevance to misfolding-induced proteostasis. Math Biosc Eng.(2020), 17(3):2470-2493. doi: 10.3934/mbe.2020135 PMID: 32233549 | |